Visual Comparation of Different Transformation Methods for Cytometry Data
Chen Hao posted on 25 Jul 2016Introduction
In biotechnology, flow cytometry is a laser- or impedance-based, biophysical technology employed in cell counting, cell sorting, biomarker detection and protein engineering, by suspending cells in a stream of fluid and passing them by an electronic detection apparatus. It allows simultaneous multiparametric analysis of the physical and chemical characteristics of up to thousands of particles per second. [1]
Flow cytometry measurements can vary over several orders of magnitude, cell populations can have variances that depend on their mean fluorescence intensities, and may exhibit heavily-skewed distributions. Consequently, the choice of data transformation can influence the output of subsequent analysis. An appropriate data transformation aids in data visualization and gating of cell populations across the range of data. [2]
BioConductor flowCore which is the most bacis R pacakge for parsing FCM data. It provides many different transfromations including linearTransform, lnTransform, logTransform, quadraticTransform, arcsinhTransform, biexponentialTransform, logicleTransform.
As stated in [3], The goal of visualizing data of a measured parameter is to determine the actual probability function (APF) of the property being investigated. However the APF is never known, the selection of a proper transformation and parameters tuning is mainly determined by visual checking of the distribution to see if the populations are clearly segerated.
For this post, the seven transformation methods provided in flowCore are explored, parameters for each transformation are slightly investigated to gain a rough understanding of their theories and the corresponding effects. At the end, a shiny APP is provided to visually and interactively compare the two most popular transformations (logicle and arcsin) for a given FCS dataset.
Loading the raw data of a FCS file
library(flowCore)
library(ggplot2)
file <- "~/Ranalysis/Sanofi Day 0 Myeloid panel_SNF002 D0.fcs"
fcs <- read.FCS(file, transformation = FALSE)## Error in read.FCS(file, transformation = FALSE): '~/Ranalysis/Sanofi Day 0 Myeloid panel_SNF002 D0.fcs' is not a valid fileSummary Information of the FCS data
fs <- pData(fcs@parameters)## Error in pData(fcs@parameters): object 'fcs' not foundfs$minExprs <- apply(fcs@exprs, 2, min)## Error in apply(fcs@exprs, 2, min): object 'fcs' not foundfs$maxExprs <- apply(fcs@exprs, 2, max)## Error in apply(fcs@exprs, 2, max): object 'fcs' not foundfs$range <- NULL## Error in fs$range <- NULL: object 'fs' not foundfs## Error in eval(expr, envir, enclos): object 'fs' not foundLinear Transformation
The formula for linear transformation is x <- a*x+b, and the crrosponding funciton is linearTransform(transformationId="defaultLinearTransform", a = 1, b = 0).
Transformation function
ggplot(data.frame(x=c(-5,5)), aes(x)) +
stat_function(fun=function(x) 1*x+0, geom="line", aes(colour="a=1,b=0")) +
stat_function(fun=function(x) 2*x+0, geom="line", aes(colour="a=2,b=0")) +
stat_function(fun=function(x) 2*x+5, geom="line", aes(colour="a=2,b=5")) +
scale_colour_manual(name="a*x+b", values=c("a=1,b=0"="blue","a=2,b=0"="red", "a=2,b=5"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20") + coord_fixed()Example
linearTrans <- linearTransform(transformationId="Linear-transformation", a=0.00001, b=1)
lt_fcs <- transform(fcs, transformList('FSC-H' ,linearTrans))## Error in transform(fcs, transformList("FSC-H", linearTrans)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'FSC-H']), main = "Before Transformation")## Error in density(fcs@exprs[, "FSC-H"]): object 'fcs' not foundplot(density(lt_fcs@exprs[ ,'FSC-H']), main = "After Transformation")## Error in density(lt_fcs@exprs[, "FSC-H"]): object 'lt_fcs' not foundNatural Logarithm Transformation (ln transformation)
The formula for ln transformation if x<-log(x)*(r/d), and the crrosponding function is lnTransform(transformationId="defaultLnTransform", r=1, d=1).
Transformation function
ggplot(data.frame(x=c(1e-20,10)), aes(x)) +
stat_function(fun=function(x) log(x)*(1/1), geom="line", aes(colour="r=1,d=1")) +
stat_function(fun=function(x) log(x)*(2/1), geom="line", aes(colour="r=2,d=1")) +
stat_function(fun=function(x) log(x)*(1/2), geom="line", aes(colour="r=1,d=2")) +
scale_colour_manual(name="log(x)*(r/d)", values=c("r=1,d=1"="blue","r=2,d=1"="red", "r=1,d=2"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20")Example
lnTrans <- lnTransform(transformationId="ln-transformation", r=1, d=1)
ln_fcs <- transform(fcs, transformList('FSC-H', lnTrans))## Error in transform(fcs, transformList("FSC-H", lnTrans)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'FSC-H']), main = "Before Transformation")## Error in density(fcs@exprs[, "FSC-H"]): object 'fcs' not foundplot(density(ln_fcs@exprs[ ,'FSC-H']), main = "After Transformation")## Error in density(ln_fcs@exprs[, "FSC-H"]): object 'ln_fcs' not foundLogarithmic Transformation
The formula for logarithmic transformation if x<-log(x,logbase)*(r/d), and the crrosponding function is logTransform(transformationId="defaultLogTransform", logbase=10, r=1, d=1). Compared with ln transformation, you can specify the base with logbase.
Transformation function
ggplot(data.frame(x=c(1e-20,1000)), aes(x)) +
stat_function(fun=function(x) log(x,exp(1))*(1/1), geom="line", aes(colour="logbase=e")) +
stat_function(fun=function(x) log(x,10)*(1/1), geom="line", aes(colour="logbase=10")) +
stat_function(fun=function(x) log(x,20)*(1/1), geom="line", aes(colour="logbase=20")) +
scale_colour_manual(name="log(x,logbase)", values=c("logbase=e"="blue","logbase=10"="red", "logbase=20"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20")Example
logTrans <- logTransform(transformationId="log10-transformation", logbase=10, r=1, d=1)
log_fcs <- transform(fcs, transformList('FSC-H', logTrans))## Error in transform(fcs, transformList("FSC-H", logTrans)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'FSC-H']), main = "Before Transformation")## Error in density(fcs@exprs[, "FSC-H"]): object 'fcs' not foundplot(density(log_fcs@exprs[ ,'FSC-H']), main = "After Transformation")## Error in density(log_fcs@exprs[, "FSC-H"]): object 'log_fcs' not foundQuadratic Transformation
The formula for quadratic transformation if x <- a*x^2 + b*x + c, and the crrosponding function is quadraticTransform(transformationId="defaultQuadraticTransform", a = 1, b = 1, c = 0).
Transformation function
ggplot(data.frame(x=c(-10,10)), aes(x)) +
stat_function(fun=function(x) 1*x^2 + 0*x + 0, geom="line", aes(colour="a=1,b=0,c=0")) +
stat_function(fun=function(x) -1*x^2 + 0*x + 0, geom="line", aes(colour="a=-1,b=0,c=0")) +
stat_function(fun=function(x) 1*x^2 + -10*x + 25, geom="line", aes(colour="a=1,b=-10,c=25")) +
stat_function(fun=function(x) 4*x^2 + 0*x + 100, geom="line", aes(colour="a=4,b=0,c=100")) +
scale_colour_manual(name="a*x^2 + b*x + c", values=c("a=1,b=0,c=0"="blue", "a=-1,b=0,c=0"="purple", "a=1,b=-10,c=25"="red", "a=4,b=0,c=100"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20")Example
quadTrans <- quadraticTransform(transformationId="Quadratic-transformation", a=1, b=1, c=0)
qd_fcs <- transform(fcs, transformList('FSC-H', quadTrans))## Error in transform(fcs, transformList("FSC-H", quadTrans)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'FSC-H']), main = "Before Transformation")## Error in density(fcs@exprs[, "FSC-H"]): object 'fcs' not foundplot(density(qd_fcs@exprs[ ,'FSC-H']), main = "After Transformation")## Error in density(qd_fcs@exprs[, "FSC-H"]): object 'qd_fcs' not foundHyperbolic arc-sine Transformation
The formula for arcsinh transformation if x<-asinh(a+b*x)+c where asinh <- function(x) {log(x + sqrt(x^2 + 1))}, and the crrosponding function is arcsinhTransform(transformationId="defaultArcsinhTransform", a=1, b=1, c=0).
Transformation function
ggplot(data.frame(x=c(-10,10)), aes(x)) +
stat_function(fun=function(x) asinh(0+1*x)+0 , geom="line", aes(colour="a=0,b=1,c=0")) +
stat_function(fun=function(x) asinh(0+4*x)+0, geom="line", aes(colour="a=0,b=4,c=0")) +
stat_function(fun=function(x) asinh(5+1*x)+5, geom="line", aes(colour="a=5,b=1,c=5")) +
stat_function(fun=function(x) asinh(0+-1*x)+0, geom="line", aes(colour="a=0,b=-1,c=0")) +
scale_colour_manual(name="asinh(a+b*x)+c", values=c("a=0,b=1,c=0"="blue", "a=0,b=4,c=0"="purple", "a=5,b=1,c=5"="red", "a=0,b=-1,c=0"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20")Example
asinhTrans <- arcsinhTransform(transformationId="ln-transformation", a=1, b=1, c=1)
as_fcs <- transform(fcs, transformList('FSC-H', asinhTrans))## Error in transform(fcs, transformList("FSC-H", asinhTrans)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'FSC-H']), main = "Before Transformation")## Error in density(fcs@exprs[, "FSC-H"]): object 'fcs' not foundplot(density(as_fcs@exprs[ ,'FSC-H']), main = "After Transformation")## Error in density(as_fcs@exprs[, "FSC-H"]): object 'as_fcs' not foundBiexponential Transformation
Biexponential Transformation is an over-parameterized inverse of the hyperbolic sine, the formula to be inverted takes the form f(x) = a*exp(b*(x-w))-c*exp(-d*(x-w))+f , and the crrosponding function is biexponentialTransform(transformationId="defaultBiexponentialTransform", a = 0.5, b = 1, c = 0.5, d = 1, f = 0, w = 0, tol = .Machine$double.eps^0.25, maxit = as.integer(5000))
Transformation function
biexp <- function(x, a = 0.5, b = 1, c = 0.5, d = 1, f = 0, w = 0,
tol = .Machine$double.eps^0.25,
maxit = as.integer(5000)){
flowCore:::biexponential_transform(input = x,
A = a,
B = b,
C = c,
D = d,
F = f,
W = w,
Tol = tol,
MaxIt = maxit)
}
ggplot(data.frame(x=c(-5000,5000)), aes(x)) +
stat_function(fun=function(x) biexp(x, a = 0.5, b = 1, c = 1, d = 1, f = 0, w = 0), geom="line", aes(colour="a=0.5,b=1,c=1,d=1,f=0,w=0")) +
stat_function(fun=function(x) biexp(x, a = 0.5, b = 2, c = 10, d = 1, f = 0, w = 0), geom="line", aes(colour="a=0.5,b=2,c=10,d=1,f=0,w=0")) +
stat_function(fun=function(x) biexp(x, a = 1, b = 1, c = 0.5, d = 1, f = 0, w = 0), geom="line", aes(colour="a=1,b=1,c=0.5,d=1,f=0,w=0")) +
stat_function(fun=function(x) biexp(x, a = 0.5, b = 1, c = 1000, d = 1, f = 0, w = 0), geom="line", aes(colour="a=0.5,b=1,c=1000,d=1,f=0,w=0")) +
scale_colour_manual(name="biexponential",
values=c("a=0.5,b=1,c=1,d=1,f=0,w=0"="blue",
"a=0.5,b=2,c=10,d=1,f=0,w=0"="purple",
"a=1,b=1,c=0.5,d=1,f=0,w=0"="red",
"a=0.5,b=1,c=1000,d=1,f=0,w=0"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20")Example
biexp <- biexponentialTransform("myTransform")
biexp_fcs <- transform(fcs, transformList('FSC-H', biexp))## Error in transform(fcs, transformList("FSC-H", biexp)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'FSC-H']), main = "Before Transformation")## Error in density(fcs@exprs[, "FSC-H"]): object 'fcs' not foundplot(density(biexp_fcs@exprs[ ,'FSC-H']), main = "After Transformation")## Error in density(biexp_fcs@exprs[, "FSC-H"]): object 'biexp_fcs' not foundLogicle Transformation
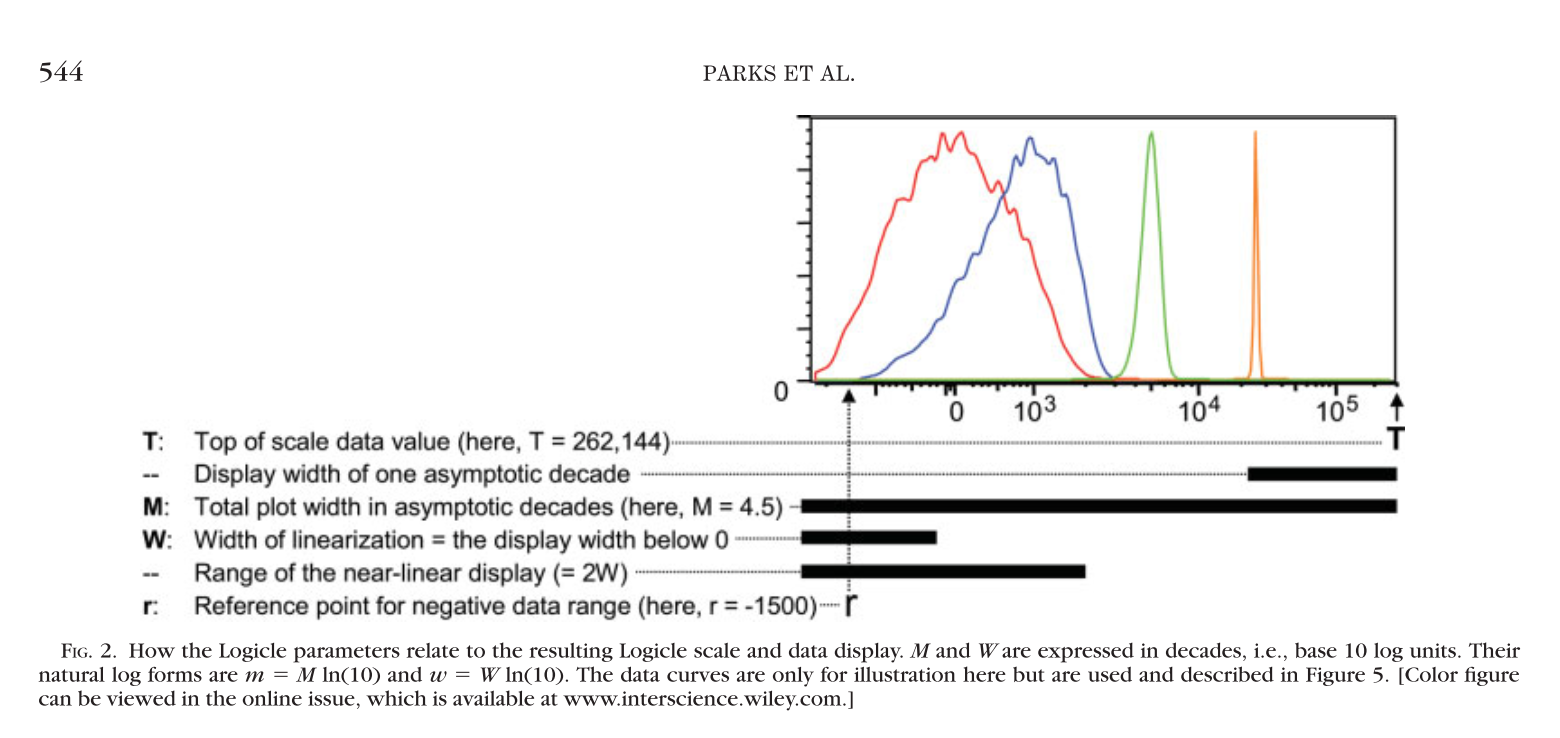
Logicle transformation creates a subset of hyperbolic sine transformation functions that provides several advantages over linear/log transformations for display of flow cytometry data.
and the crrosponding function is logicleTransform(transformationId="defaultLogicleTransform", w = 0.5, t = 262144, m = 4.5, a = 0)
Transformation function
lgcl <- function(x, w = 0.5, t = 262144, m = 4.5, a = 0){
flowCore:::logicle_transform(input = x,
T = t,
W = w,
M = m,
A = a,
isInverse = FALSE)
}
ggplot(data.frame(x=c(-5000,5000)), aes(x)) +
stat_function(fun=function(x) lgcl(x, w = 0.5, t = 5000, m = 4.5, a = 1), geom="line", aes(colour="w=0.5,t=5000,m=4.5,a=1")) +
stat_function(fun=function(x) lgcl(x, w = 0.5, t = 2500, m = 4.5, a = 0), geom="line", aes(colour="w=0.5,t=2500,m=4.5,a=0")) +
stat_function(fun=function(x) lgcl(x, w = 0.5, t = 5000, m = 3, a = 0), geom="line", aes(colour="w=0.5,t=5000,m=3,a=0")) +
stat_function(fun=function(x) lgcl(x, w = 1, t = 5000, m = 4.5, a = 0), geom="line", aes(colour="w=1,t=5000,m=4.5,a=0")) +
scale_colour_manual(name="logicle",
values=c("w=0.5,t=5000,m=4.5,a=1"="blue",
"w=0.5,t=2500,m=4.5,a=0"="purple",
"w=0.5,t=5000,m=3,a=0"="red",
"w=1,t=5000,m=4.5,a=0"="green")) +
theme_bw() + geom_hline(yintercept = 0, colour = "gray20") + geom_vline(xintercept = 0, colour = "gray20")We can see that m controls the range of positive values, w controls the range of negative values,
a changes the slope, t is the maximal x value corresponds to m. A nice explanation is included in [4].
Example
lgcl <- logicleTransform("myTransform", w = 0.5, t= 262144, m = 4.5)
lgcl_fcs <- transform(fcs, transformList("PE-A", lgcl))## Error in transform(fcs, transformList("PE-A", lgcl)): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'PE-A']), main = "Before Transformation")## Error in density(fcs@exprs[, "PE-A"]): object 'fcs' not foundplot(density(lgcl_fcs@exprs[ ,'PE-A']), main = "After Transformation")## Error in density(lgcl_fcs@exprs[, "PE-A"]): object 'lgcl_fcs' not foundAnd in flowCore, there is a funciton estimateLogicle which can help automatic determine the parameters for logicleTransform.
And it works as below:
# flowCore:::.lgclTrans
function (dat, p, t, m, a = 0, q = 0.05, type = "instrument")
{
type <- match.arg(type, c("instrument", "data"))
transId <- paste(p, "logicleTransform", sep = "_")
rng <- range(dat)
dat <- exprs(dat)[, p]
if (missing(t)) {
if (type == "instrument")
t <- rng[, p][2]
else t <- max(dat)
}
if (missing(m)) {
if (type == "instrument")
m <- 4.5
else m <- log10(t) + 1
}
dat <- dat[dat < 0]
w <- 0
if (length(dat)) {
r <- .Machine$double.eps + quantile(dat, q)
w = (m - log10(t/abs(r)))/2
if (w < 0)
stop("w is negative!Try to increase 'm'")
}
logicleTransform(transformationId = transId, w = w, t = t,
m = m, a = a)
}algcl <- estimateLogicle(fcs, channels = c('PE-A'))## Error in estimateLogicle(fcs, channels = c("PE-A")): object 'fcs' not foundalgcl_fcs <- transform(fcs, algcl)## Error in transform(fcs, algcl): object 'fcs' not foundpar(mfrow=c(1,2))
plot(density(fcs@exprs[ ,'PE-A']), main = "Before Transformation")## Error in density(fcs@exprs[, "PE-A"]): object 'fcs' not foundplot(density(algcl_fcs@exprs[ ,'PE-A']), main = "After Transformation")## Error in density(algcl_fcs@exprs[, "PE-A"]): object 'algcl_fcs' not foundShiny APP
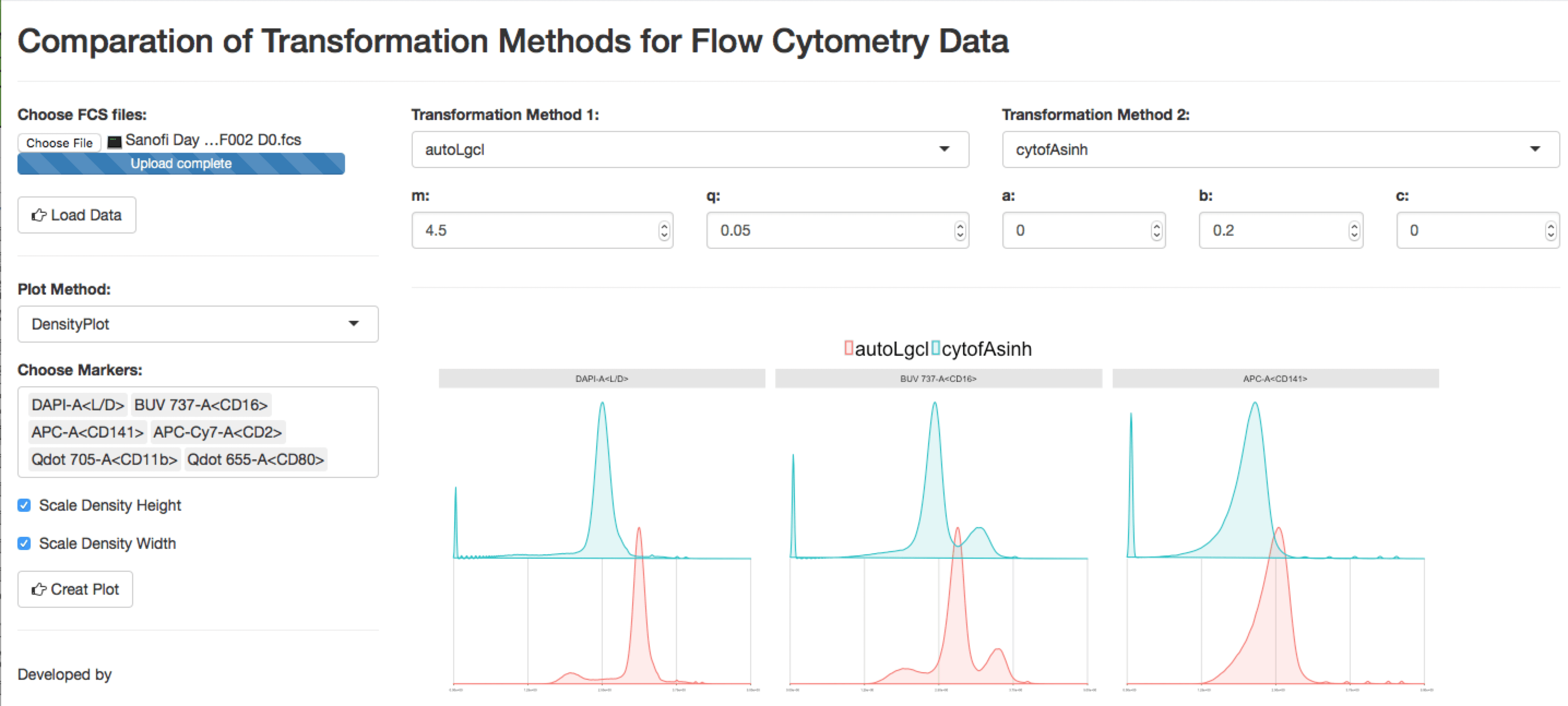
In our cytofkit pacakge, we provide another two customized transfomations: autoLgcl (modified from logicle) and cytofAsinh (modified from arcsinh).
#' Noise reduced arsinh transformation
#'
#' Inverse hyperbolic sine transformation (arsinh) with a cofactor of 5, reduce noise from negative values
#' Adopted from Plos Comp reviewer
#'
#' @param value A vector of numeric values.
#' @param cofactor Cofactor for asinh transformation, default 5 for mass cytometry data.
#' @noRd
#' @return transformed value
cytofAsinh <- function(value, cofactor = 5) {
value <- value-1
loID <- which(value < 0)
if(length(loID) > 0)
value[loID] <- rnorm(length(loID), mean = 0, sd = 0.01)
value <- value / cofactor
value <- asinh(value) # value <- log(value + sqrt(value^2 + 1))
return(value)
}
#' a modified version of "estimateLogicle" from flowCore
#'
#' Used boxplot outlier detection to filter outliers in negative values
#' before calculating the r using the fifth percnetile of the negative values.
#'
#' @param x A flowFrame object.
#' @param channels Channel names to be transformed.
#' @param m The full width of the transformed display in asymptotic decades. m should be greater than zero.
#' @param q The percentile of negative values used as reference poiont of negative range.
#' @importFrom methods is
#' @importFrom flowCore logicleTransform
#' @noRd
#' @return a list of autoLgcl transformations
autoLgcl <- function(x, channels, m = 4.5, q = 0.05) {
if (!is(x, "flowFrame"))
stop("x has to be an object of class \"flowFrame\"")
if (missing(channels))
stop("Please specify the channels to be logicle transformed")
indx <- channels %in% colnames(x@exprs)
if (!all(indx))
stop(paste("Channels", channels[!indx], "were not found in the FCS file.\n ",
sep = " "))
trans <- lapply(channels, function(p) {
data <- x@exprs[, p]
w <- 0
t <- max(data)
ndata <- data[data < 0]
## use 1.5 * IQR to filter outliers in negative values
nThres <- quantile(ndata, 0.25) - 1.5 * IQR(ndata)
ndata <- ndata[ndata >= nThres]
transId <- paste(p, "autolgclTransform", sep = "_")
if (length(ndata)) {
r <- .Machine$double.eps + quantile(ndata, q)
## Check to avoid failure of negative w
if (10^m * abs(r) <= t) {
w <- 0
} else {
w <- (m - log10(t/abs(r)))/2
if(w>2) {
w <- 0.5
}
}
}
logicleTransform(transformationId = transId,
w = w, t = t, m = m, a = 0)
})
transformList(channels, trans)
}To compare the transformation methods (autoLgcl, logicle, cytofAsinh, arcsinh) for a given FCS file, a shiny APP is build and provided with link https://chenhao.shinyapps.io/TransformationComparation_shinyAPP/. A screenshot of the app is as below:
Reference
[1] From wikipedia Flow cytometry
[2] Finak G, Perez J, Weng A, Gottardo R. Optimizing transformations for automated, high throughput analysis of flow cytometry data. BMC Bioinformatics. 2010;11: 546. doi:10.1186/1471-2105-11-546.
[3] Berg RA Van Den, Hoefsloot HCJ, Westerhuis J a, Smilde AK, Werf MJ Van Der, van den Berg R a, et al. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7: 142. doi:10.1186/1471-2164-7-142.
[4] Parks DR, Roederer M, Moore WA. A new “logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytom Part A. 2006;69: 541–551.
Session Information
sessionInfo()## R version 3.3.2 (2016-10-31)
## Platform: x86_64-apple-darwin13.4.0 (64-bit)
## Running under: macOS Sierra 10.12.3
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] flowCore_1.40.3 ggplot2_2.2.1 wordcloud_2.5
## [4] RColorBrewer_1.1-2 dplyr_0.5.0 knitr_1.15.1
##
## loaded via a namespace (and not attached):
## [1] graph_1.52.0 Rcpp_0.12.9 cluster_2.0.5
## [4] magrittr_1.5 BiocGenerics_0.20.0 munsell_0.4.3
## [7] lattice_0.20-34 colorspace_1.3-2 rrcov_1.4-3
## [10] R6_2.2.0 pcaPP_1.9-61 stringr_1.1.0
## [13] highr_0.6 plyr_1.8.4 tools_3.3.2
## [16] parallel_3.3.2 grid_3.3.2 Biobase_2.34.0
## [19] gtable_0.2.0 DBI_0.5-1 corpcor_1.6.8
## [22] matrixStats_0.51.0 digest_0.6.12 lazyeval_0.2.0
## [25] assertthat_0.1 tibble_1.2 robustbase_0.92-7
## [28] evaluate_0.10 slam_0.1-40 labeling_0.3
## [31] stringi_1.1.2 DEoptimR_1.0-8 scales_0.4.1
## [34] stats4_3.3.2 mvtnorm_1.0-5